
PROBER
Tutorial
Section 4
PROBER: the main interface for probe design
Close
the Tolerance & MerMatch window and return to the main PROBER
graphical interace
First
load in the original (*.fa or *.dna) DNA sequence (from DAS.DNA) by clicking
File > Load DNA Sequence
The loaded DNA sequence will appear at the bottom of the program.
Now load the masked (*.mnx1
or *.mnx2) DNA
sequence (from MerMatch & Tolerance) by clicking File > Load MNX
Sequence.
The
loaded MNX sequence will appear at the bottom of the program.
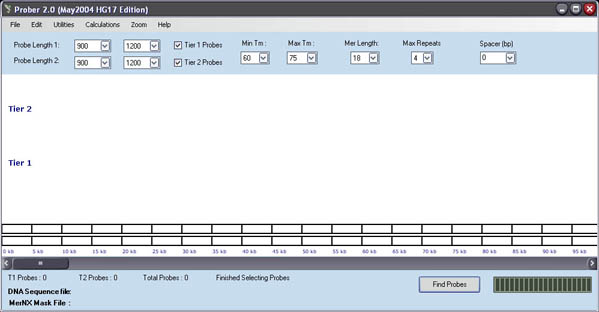
![]()
A number of parameters appear in the main
PROBER window.
Tier 1 Probe Length : This is the size range in Bp of the final DNA probes,
Tier 1 should have a small range.
Tier
2 Probe Length : The size range of the Tier 2 probes should be more relaxed
then Tier 1 and have a larger range
Min
Tm : This is the minimum primer melting temperature in celsius that is
allowed during probe selection (def = 60 C)
Max
Tm : This is the maximum primer melting temperature in celsius that is
allowed during probe selection (def = 75 C)
Max
Repeats : The number of consecutive polynucleotides allowed in the primers
Spacer:
This is the number of nucleotides that will be inserted between probes,
generally this is set to 0
Set
the following parameters for probe selection:
Tier 1 Probe Length : 900 - 1100
Tier
2 Probe Length : 1000 - 1400
Min
Tm :
60 C
Max
Tm :
70 C
Repeat
Nucleotides :
4
MerMatch
Length:
18

Once
the parameters for probe selection are set, click the button in the lower
right hand corner - 'Find Probes'
Prober
will run for approximately 1 minute, Please proceed to the next section