
INTRODUCTION
PROBER
is useful in designing DNA probes for determining genomic copy number
in cells.
Genomic DNA sequences are retrieved from a server, masked for repetitive
exact string matches in the human genome and analyzed for contiguously
repeat free regions of sufficient aggregate length. These regions are
then searched for optimized PCR forward and reverse primers, resulting
in a collection of oligonucleotide probes. These probes are then be PCR
amplified and purified, resulting in a collection of probes that we combine
into a cocktail for FISH analysis.
Three easy steps are taken to create highly specific DNA probes. They
are described in the order in which they are employed by the software
application. The only requirement from the user are genomic start and
stop coordinates and a human genome freeze.
PROGRAMS
DAS.DNA
Retrieves a human genomic DNA sequence 10-100kb in length from a Distributed
Annotation Sever (DAS) server given coordinates and a freeze.
MER-MATCH
Masks repetitive areas of the DNA sequence with 'N' using a mermatch length
with exact string matching to a mer dictionary genome database. In other
words, a word length is matched against the human genome to identify repetitive
elements in the DNA sequence for probe design.
TOLERANCE
Next, MerMatch will find the largest regions of contiguous good sequence
based on cumulative sums and mark these regions with integers based on
their size. The largest contigous DNA sequence that is repeat free is
ranked with the lowest integer and the collection of regions are ordered
for subsequent probe selection.
PROBER
Selects
probes of 100-2000bp from the highest ranked non-repetitive DNA sequence
first using distance matricies. PROBER scores all possible permutations
of primers within a size range (15-30bp) and follow primer design guidelines
to eliminate 'bad' primers. The
optimal primers are selected based on probe size and matching melting
temperatures. The final output includes a full report with a number of
probe statistics and a short report containing only the forward and reverse
primer sequences that may be used for ordering primers.
OUTPUT
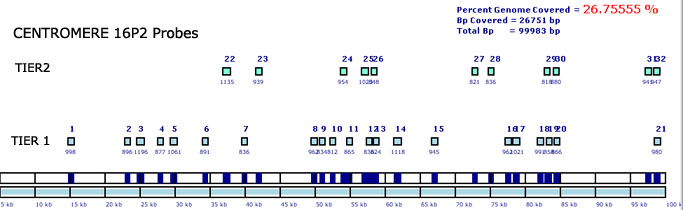
The software outputs an image containing the location of each probe relative
to the 20-100kb sequence. Two rounds of probe selection are indicated
by 'Tiers' with minimal overlap in the genomic sequence that they cover.
Repetitive areas are completely avoided for primer selection. The probes
are identified by their ID on the upper portion of each rectangle and
the probe length underneath. A percentage of genome coverage is also indicated
in the upper right hand corner. If the PGC% is to low (for example <
20%) the probe selection can be repeated with less stringent parameters.
A text report can be saved containing specific information on each probe
and primer pair including Primer length, Tm, Primer Sequence, Probe Sequence
and Probe Length. In addition, a short report containing only the forward
and reverse primer sequences in a seperate file can be saved to facillitate
the ordering process.
::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
-TIER-1-PROBES:::::::[FULL PROBE REPORT]::::::::::::::::::::
::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::::
Probe
ID 1
PrimerFseq :CTGTGCCGTTAACTCGAATG
PrimerRseq :GCACTCTCGAATGCGCCG
PrimerFseq Unmasked :CTGTGCCGTTAACTCGAATG
PrimerRseq Unmasked :CGGCGCATTCGAGAGTGC
PrimerF_length :20
PrimerR_length :18
PrimerF_loc :14758
PrimerR_loc :15756
PrimerF_tm :60
PrimerR_tm :60
Probe Length : 1016
Probe Sequence :
CTGTGCCGTTAACTCGAATGCTAGCCTGGTTAGGCGGGATTTCTCTGCTGGTAGAAATATGCCTTCTTCC
GTCTGCAAATTCCTGCAGCATCAGACAAACCCAGCCTACAATATTTAGACTGATTTTTACCAACTTAAT
GGCAGGGTCCATTTTGAAGGGGAAGCCAACAGTGTCTATGAGAGACGGGGTGGGGACGTTGGTGAA
AGAGAGGGGCTGAGGGTGGGTGAGTCGTATGTGCATCAAGTTTGGCCTCCAGTGAATCTTGGTCTAGA
ATGCTAGAGCCAGTGCTCCCACCCCTCAGTGCCCTGTCTGCTTCCTTACTGAGCAGGCTTGTAGGGCTTC
GTGAGATGAGGAGTCCCCTCCCCGTAGGGCTGCTGAGGGCTGAATGTGAAGTTCTTGGGTGGCCTGTG
TGGCTGGGATGAGTGGGCAT
Please visit the 'tutorial' section to
go through the steps required for designing oligo probes for a sample
locus