
PROBER
Tutorial
Section 3
Tolerance : find unique genomic blocks for probe selection
Continue to the lower portion of the same window titled ' Tolerance '
After the masked DNA sequence appears in the upper window, the controls for Tolerance become active.
The "min.length" is the number of consecutive n-mers with multiple exact matches above a threshold k. The "length" is the minimum size of amplifiable region on which to search for genomic blocks. "Repeat.tolerance" is how many mers with multiple exact matches above threshold k that are allowed, consecutive or not. At 0.8 80% of mers are allowed. Adjusting "min.length" and "repeat.tolerance" will affect the stringency and size of the genomic blocks of unique sequence that the tolerance algorithm finds.
For this example use the following (default) parameters:
min.length: 10
length: 100
repeat.tolerance: 0.8
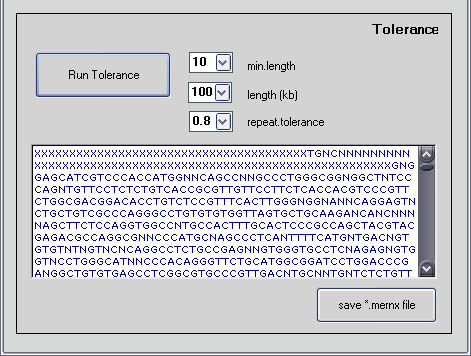
Press 'run.amp.coords' (the application should run for < 1 min)
The output of find.amp.coords is a *.mnx file. This file contains XXX's to mark the interstitial regions of unique genomic blocks for probe and primer selection.
Press 'save *.mernx file'
(Name the file *.mnx1 to note that the "mer.count.cutoff" was 1 or *.mnx2 to denote that the cutoff was 2)
Save the file in the same folder as the DNA sequence from DAS.DNA.
Close the MerMatch / Tolerance Window.