
SIMULATIONS
Avoiding Repetitive DNA Elements
In
order to test the ability of the genomic block algorithm in avoiding repetitive
DNA elements that occur in the human genome, a series of simulations were
conducted using inserted LINEs and LTRs. A highly unique 100kb genomic
sequence was obtained from the human genome (chr7:28,300,000-28,400,000bp)
as a control sequence, to which LINEs and LTRs were inserted at regular
intervals.

Control
Sequence from UCSC HG17| chr7:28,300,000-28,400,000bp with very low
repeats

Control Sequence tiled with 2 tiers of DNA probes
covering 94.57% of the locus (Unique 100kb Genomic Sequence)
Insertion
of LINEs
21% of the human genome consists of Long Interspersed Elements (LINES)
that range in size from a few hundred to 9000 bp in size. Most LINEs are
artifacts that are no longer mobile in the human genome, however, functional
LINEs such as L1 elements encode 3 proteins including an endonuclease
that cuts DNA and a reverse transcriptase that makes a DNA copy of an
RNA transcript. For this simulation, we used a 7594 bp L1-LINE (HG17|chr7:141089122-141096716)
and inserted it at specific locations (20kb, 40kb and 60kb) in the 100kb
control sequence. All genomic blocks were identified neighboring the inserted
LINES and probes were selected from unique regions, rendering them highly
specific to this locus. In this simulation no probes intersected the regions
containing the inserted LINEs.
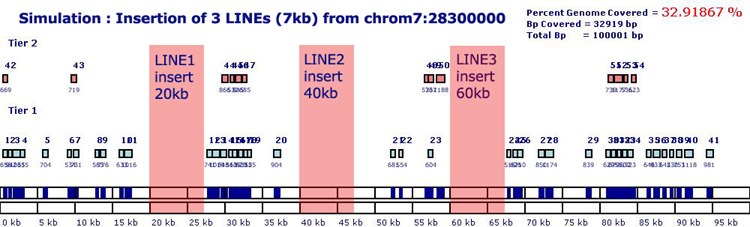
Control Sequence with 7.6kb LINEs inserted at 20kb,
40kb and 60kb. Probes were tiled across 32.91% of the locus.
Insertion
of LTRs
Long Terminal Repeat Elements (LTRs) are a class of repetitive elements
that include transposons and retrotransposons. They have been dubbed 'junk'
DNA, because they have no benefit to the host. Richard Dawkins refers
to transponsons as 'selfish' DNA, for their only function appears to be
to make copies of theirselves. Transposons have inverted repeats on either
5' / 3' end of the sequence and range in size from a few hundred bp to
10kb. For this simulation two consecutive 3363 bp LTRs (LTR1, LTR2, LTR3)
from HG17|chr7:128813516-128816879 were inserted at specific locations
into the unique control sequence. The LTRs positioned at 10kb, 50kb and
72kb were avoided by the genomic block algorithm. The primers and probes
selected from genomic blocks surrounding the LTRs are highly specific
and unqiue to the control locus.
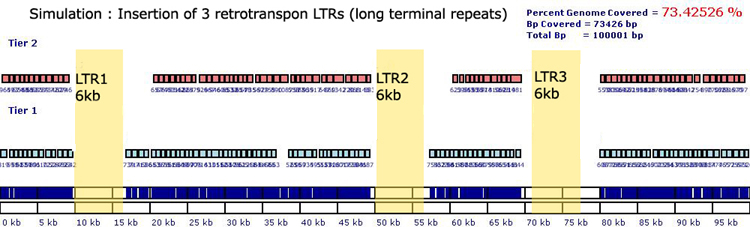
Control Sequence with 2x 3.36kb LINEs inserted at 10kb,
50kb and 72kb. Probes covered 73.42% of the locus.
Simulating the Insertion of repeatitive elements such as LTRs and LINEs
to which the genomic block algorithm was able to identify and avoid repetitve
elements, suggests that PROBER is capable of avoiding repetive
areas of the genome for probe selection.