
PROBER Tutorial
Section 1
DasDNA : retrieve a DNA sequence from a DAS server
Open the graphical interface to DAS.DNA by clicking on Utilities > DAS.DNA from the menu bar.
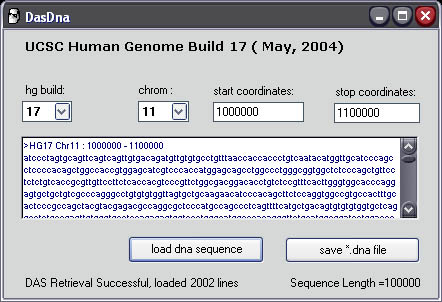
Here HG build will correspond to the version of PROBER that you downloaded. For this tutorial we downloaded and installed the Build 17 (HG17 May,2004 UCSC) version of PROBER 2.0
To retrieve a genomic sequence select the chromosome number and start/stop coordinates. If the sequence length is less then 20kb or larger then 100kb, a warning message will be displayed and the retrieval will be aborted.
Enter:
chrom: 11 start: 1000000 stop : 1100000
Press 'load dna sequence' to send a request to a Distributed Annotation Server (DAS) at UCSC and an xml file will be returned.
Note: Your computer must be online for this program to run.
The DNA sequence within the xml file will be parsed and displayed in the main window. Next, click 'save *.dna file' and save the file in a new project folder on your hard drive. At this point you should create a new folder for each probe that you create for saving all of your PROBER files. The file will be saved as a fasta (*.fa) file with a header describing the freeze and genomic location of the DNA sequence in the file.
(Alternatively, you can use a genomic DNA sequence that has been downloaded from another source in fasta format)
Close the DAS.DNA window.