
PROBER
Tutorial
Section 2
MerMatch: mask repetitive DNA sequences in the human genome
Launch the MerMatch & Tolerance window by clicking on the 'Utilities'
Menu > MerMatch / Tolerance
Press
'Load
DNA sequence' that was previously saved by DAS.DNA to load in the genomic
DNA sequence.
Note
: The PROBER folder and files must be installed in C:\PROBER.
(Otherwise MerMatch will output an error and will
not be able to run)
In
the MerMatch / Tolerance graphical interface set
the Load in RAM parameter to 1 (true) if you have 1 gig of RAM available
on your machine. If your computer has less then 1 gigabyte of RAM
then set Load in RAM to 0 (false).
Set the mer.match.length to 18 (default). This is the window size of nucleotides
that will be matched against the human genome to determine how many times
it occurs. After matching the window against the human genome,
the number of exact string matches is reported and the window is moved
up by one base pair.
This cycle continues until the number of matches for each window
within the DNA sequence is determined.
For example using a mer.match.length of 18 on a sequence such as this:
AGCTAGCATAGAGATCGACTAGCTACT
Will result in the following query strings to match against the human
genome:
AGCTAGCATAGAGATCGA
= 1
GCTAGCATAGAGATCGAC
= 2
CTAGCATAGAGATCGACT
= 4
TAGCATAGAGATCGACTA
= 1
AGCATAGAGATCGACTAG
= 1
GCATAGAGATCGACTAGC
= 1
CATAGAGATCGACTAGCT
= 5
ATAGAGATCGACTAGCTA
= 3
TAGAGATCGACTAGCTAC
= 1
AGAGATCGACTAGCTACT
= 2
Each substring has a corresponding frequency in the human genome. Set
the "mer.count.cutoff" to 1 (default). This will mask any substrings
that occur at a frequency > 1.
ANNTAGNNTNGAGATCGACTAGCTACT
Substrings that are repetive in the human genome (mer.count.cutoff >
1) are masked with 'N'.
Run MerMatch with the following (default) parameters:
Load
in RAM: 1
mer.match.length:
18
mer.count.cutoff: 3
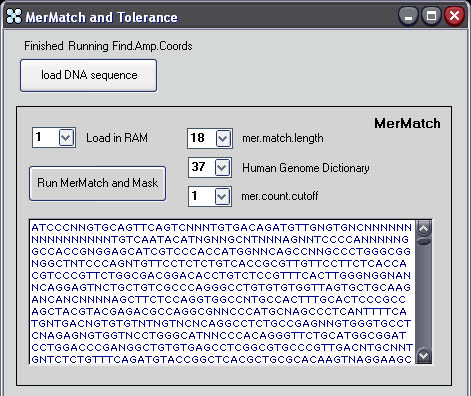
Press
'Run Mer Engine'
A
DOS window will launch the MerMatch executable. Depending on the length
of the DNA sequence, RAM and processor speed, the software will run for
several minutes. Once it is finished, the masked DNA sequence will appear
in the text window below. Do
not close this window.
Please proceed to the next section.