
New Version Released ! (V2.1)
PROBER is an oligonucleotide primer design software application that can
generate highly specific probes for use in fluorescence in-situ hybridization
(FISH) and other in-situ
labeling methods by densely tiling relatively small genomic intervals.
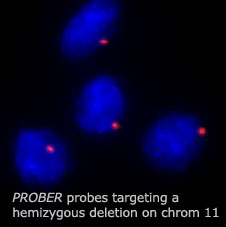 Generating
Tiling Oligonucleotide Probes (TOPs) requires software capable of masking
repetitive genomic sequences and saturating unique contiguous blocks with
small (100-2000bp), DNA probes that will generate a single, strong fluorescent
signal for regions as small as a single gene.
Generating
Tiling Oligonucleotide Probes (TOPs) requires software capable of masking
repetitive genomic sequences and saturating unique contiguous blocks with
small (100-2000bp), DNA probes that will generate a single, strong fluorescent
signal for regions as small as a single gene.
The program employs a genomic block identification algorithm that we have
designed which uses the cumulative sums of areas demarcated as repetitive
in order to identify the largest contiguous blocks of unique genomic sequence
for the target locus. A probe search algorithm was developed to find the
location of the optimal forward and reverse primers of a desired size
range within these genomic blocks for probe amplification. In order to
test the efficacy of our software, we have designed a number of probes
for genomic amplifications that have been detected by Representational
Oligonucleotide Microarray Analysis (ROMA) in breast cancer tumor samples.
We have determined that PROBER is capable of designing tiling oligo probes
for FISH with high genomic specificity both in silico and in
vitro.
Prober was written in C# 2005 and has sub-programs that are complied as
executables that were written in c, perl and splus. When running PROBER
you will need to have the .net
framework 2.0 installed on your pc machine.
Nicholas Navin
Wigler Lab
Cold Spring Harbor Laboratory
navin@cshl.edu